Study Notes
Overview
Inheritance is the cornerstone of genetics, exploring how traits are passed from one generation to the next. For Edexcel A-Level Biology, this isn't just about drawing Punnett squares; it's about precision, application, and interpretation. This topic requires you to be a biological detective, using genetic clues to predict outcomes, analyse data, and explain complex patterns. You'll delve into the mechanisms of monohybrid, dihybrid, and sex-linked crosses, and extend your understanding to non-Mendelian scenarios like autosomal linkage and epistasis. A strong grasp here is vital as it forms synoptic links with topics like meiosis, gene expression, and evolution. Examiners frequently test this area with high-mark, multi-step questions, so a solid foundation is key to success.
Key Concepts
1. Genes, Alleles, and Loci
It is absolutely critical to use terminology with precision. A gene is a sequence of DNA that codes for a functional polypeptide or RNA molecule. Its position on a chromosome is called its locus. An allele is a specific version of a gene. For example, the gene for eye colour has alleles for blue, brown, green, etc. Candidates who write 'the dominant gene' instead of 'the dominant allele' will not be awarded marks.
- Genotype: The alleles an organism possesses for a particular gene (e.g.,
AA,Aa, oraa). - Phenotype: The observable characteristics of an organism resulting from its genotype and the environment (e.g., tall plant, blue eyes).
- Dominant Allele: An allele that is always expressed in the phenotype, even if only one copy is present (e.g.,
AinAa). - Recessive Allele: An allele that is only expressed in the phenotype if two copies are present (e.g.,
ainaa). - Homozygous: Having two identical alleles for a gene (e.g.,
AAoraa). - Heterozygous: Having two different alleles for a gene (e.g.,
Aa).
2. Monohybrid Inheritance
This involves tracking the inheritance of a single gene. The classic example is a cross between two heterozygous parents (Aa x Aa), which produces a predictable 3:1 phenotypic ratio in the offspring (three dominant phenotype, one recessive phenotype) and a 1:2:1 genotypic ratio (one homozygous dominant, two heterozygous, one homozygous recessive).
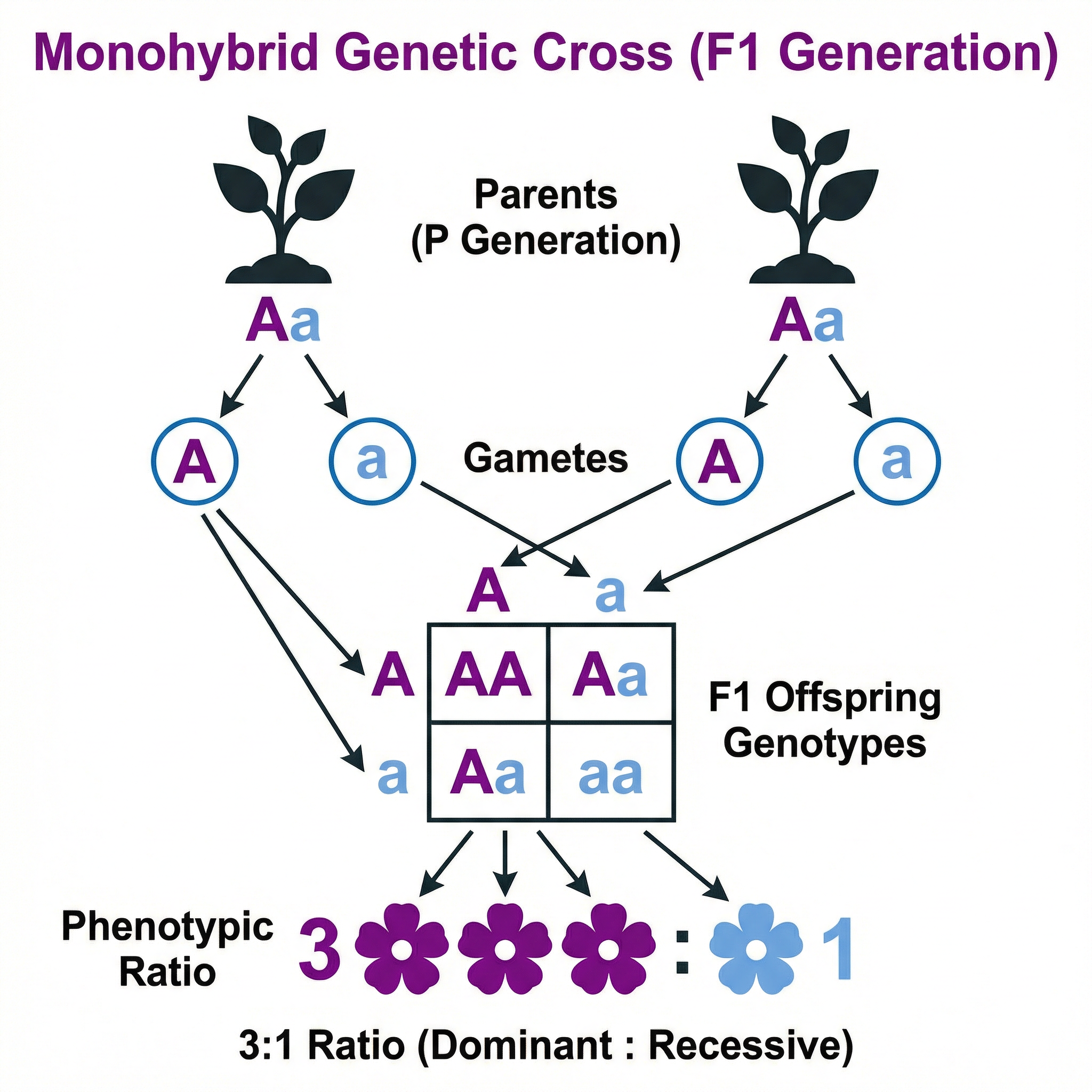
Examiner Tip: For a genetic diagram question, credit is given for:
- Stating parental phenotypes and genotypes.
- Correctly identifying the gametes (and circling them).
- A correctly constructed Punnett square.
- Stating the offspring phenotypic ratio.
3. Dihybrid Inheritance
This tracks the inheritance of two different genes, located on different chromosomes. A cross between two parents heterozygous for both genes (AaBb x AaBb) produces the classic 9:3:3:1 phenotypic ratio.
- 9/16 show both dominant traits
- 3/16 show one dominant, one recessive trait
- 3/16 show the other dominant, one recessive trait
- 1/16 show both recessive traits
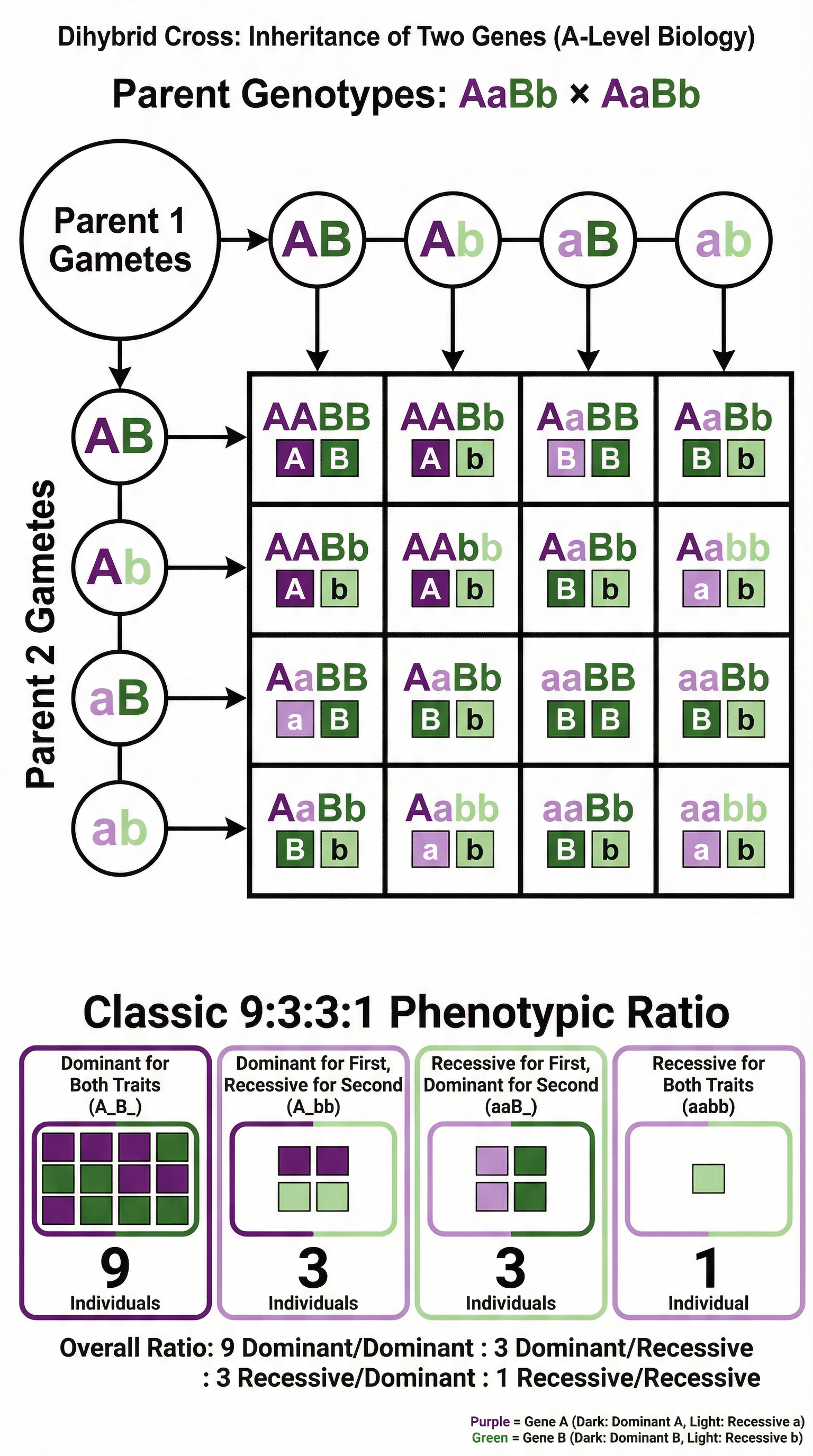
Crucial Point: This ratio only occurs when the genes are on different chromosomes (unlinked). If you observe a different ratio, it points towards other genetic interactions.
4. Non-Mendelian Inheritance: Linkage & Epistasis
Autosomal Linkage: This occurs when two or more genes are located on the same autosome (non-sex chromosome). Because they are physically linked, their alleles are inherited together as a single unit unless separated by crossing over during meiosis. This reduces the number of possible gamete combinations and skews the expected dihybrid ratio. The closer the loci of the genes, the less likely they are to be separated by crossing over, and the more they behave as a single gene.
Epistasis: This is where one gene masks or suppresses the expression of another gene at a different locus. For example, in recessive epistasis, the homozygous recessive genotype at one locus (e.g., ee) prevents the expression of alleles at a second locus. This modifies the 9:3:3:1 ratio into other forms, such as 9:3:4 or 9:7. You must be able to recognise these from data.
5. Sex Linkage
This refers to traits determined by genes located on the sex chromosomes (X or Y). Most are X-linked. Males are hemizygous for X-linked traits because they only have one X chromosome. This means a single recessive allele on their X chromosome will be expressed in the phenotype. This is why X-linked recessive disorders like colour blindness and haemophilia are far more common in males.
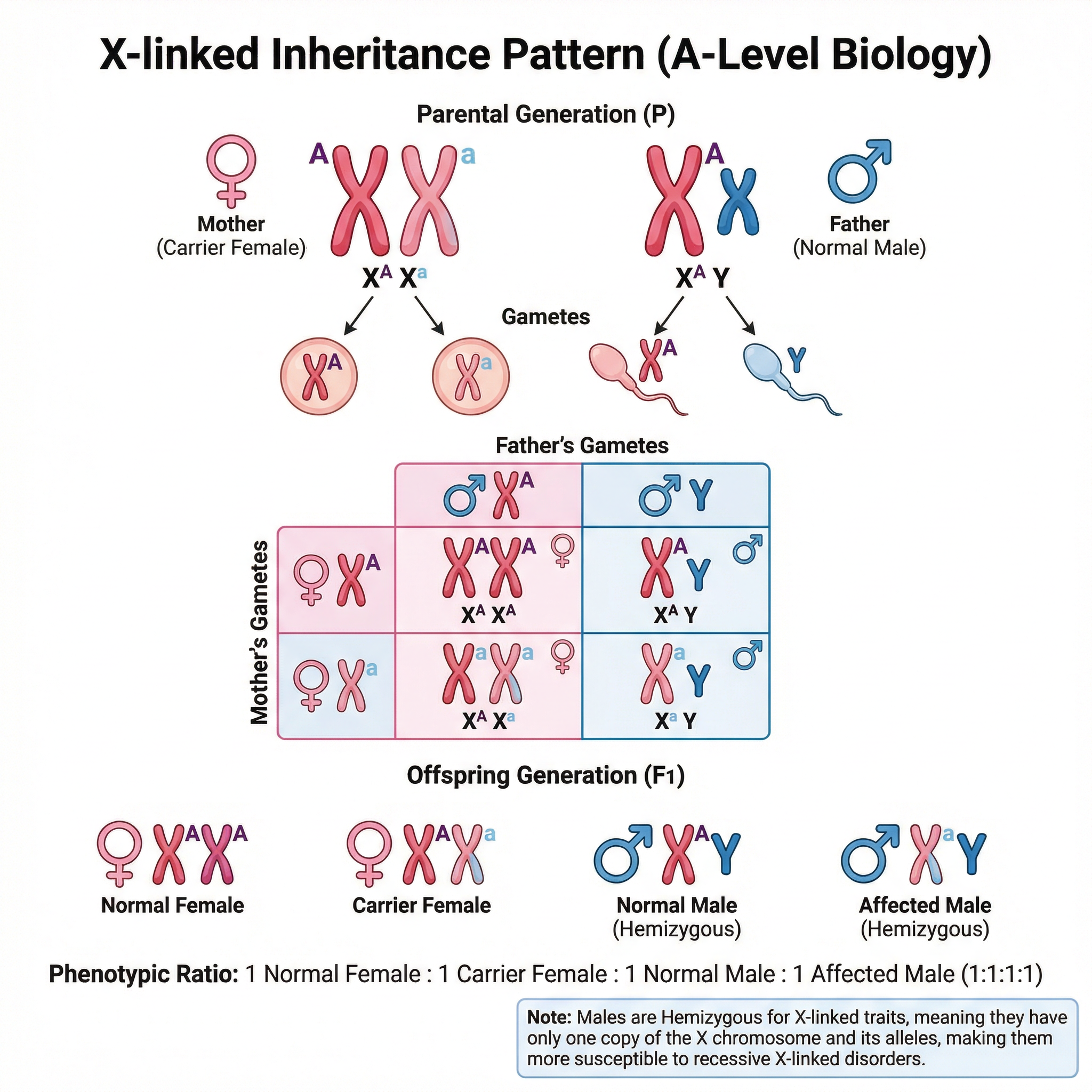
Common Mistake: Candidates often fail to show the sex chromosomes (X and Y) in diagrams or incorrectly attach alleles to the Y chromosome for X-linked traits. Full marks require clear representation like X^A, X^a, and Y.
Mathematical/Scientific Relationships
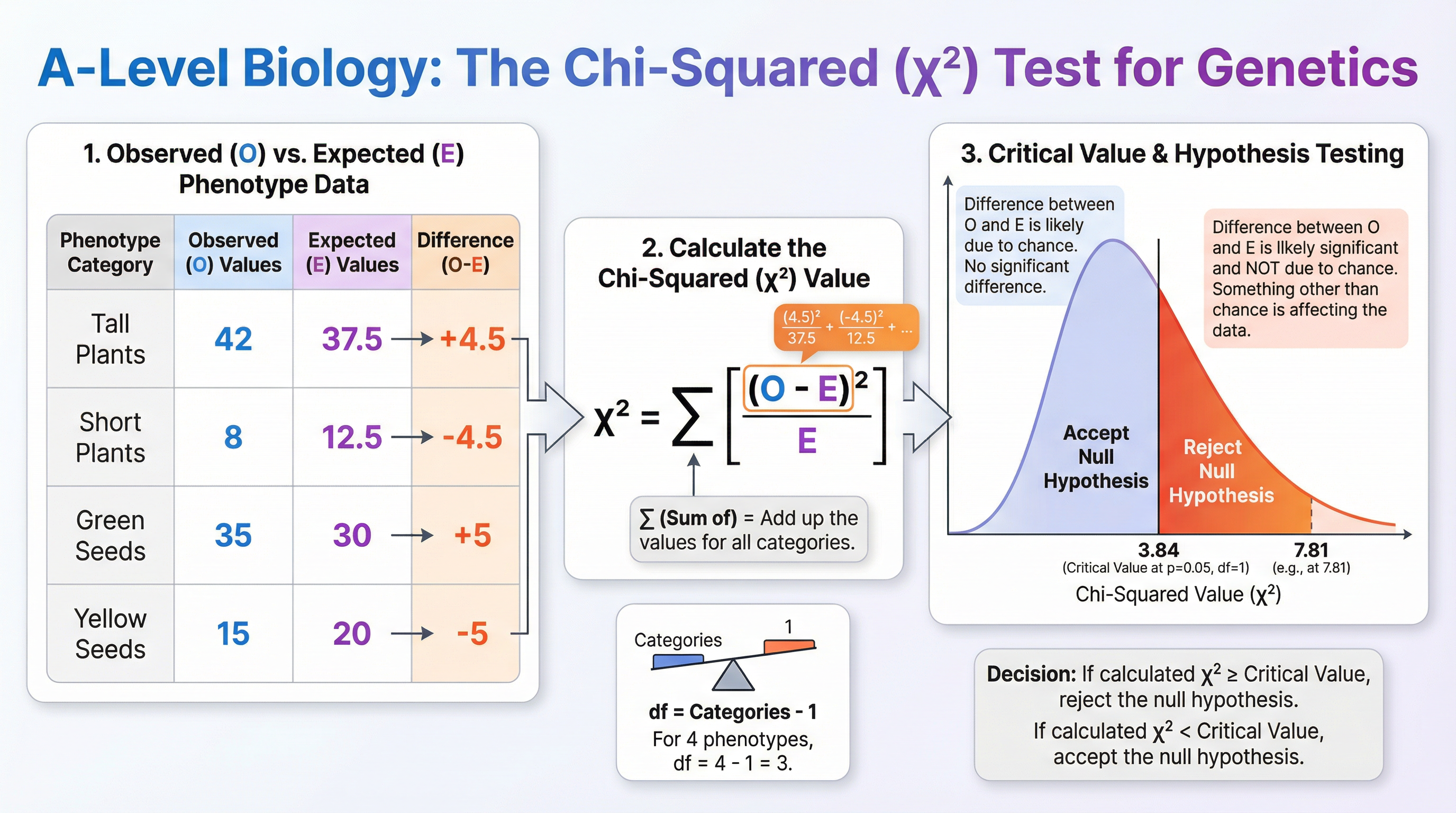
The Chi-Squared (χ²) Test
This is a statistical test used to determine if the difference between observed and expected results in a genetic cross is statistically significant or simply due to chance. It is a critical skill for AO2 and AO3 marks.
Formula: χ² = Σ [ (Observed - Expected)² / Expected ]
- Σ = Sum of
- O = Observed frequency
- E = Expected frequency
Steps:
- State the Null Hypothesis (H₀): "There is no significant difference between the observed and expected frequencies. Any difference is due to chance."
- Calculate Expected Values: Based on the relevant Mendelian ratio (e.g., 3:1, 9:3:3:1).
- Calculate the χ² value: Using the formula for each phenotypic category and summing the results.
- Determine Degrees of Freedom (df):
df = number of phenotypic categories - 1. - Compare to Critical Value: Find the critical value from a table at p=0.05 for your calculated degrees of freedom.
- If χ² < critical value: Accept H₀. The difference is not significant and is likely due to chance.
- If χ² ≥ critical value: Reject H₀. The difference is significant. A biological factor (e.g., linkage, epistasis) is likely affecting the results.
Practical Applications
Inheritance principles are the bedrock of genetic counselling, animal and plant breeding, and conservation. For example, understanding the inheritance of genetic disorders allows counsellors to advise prospective parents on the probability of their children being affected. In agriculture, breeders use knowledge of dihybrid crosses and linkage to select for desirable traits in crops and livestock, such as high yield and disease resistance. This is a required practical area, often tested through data analysis questions.
